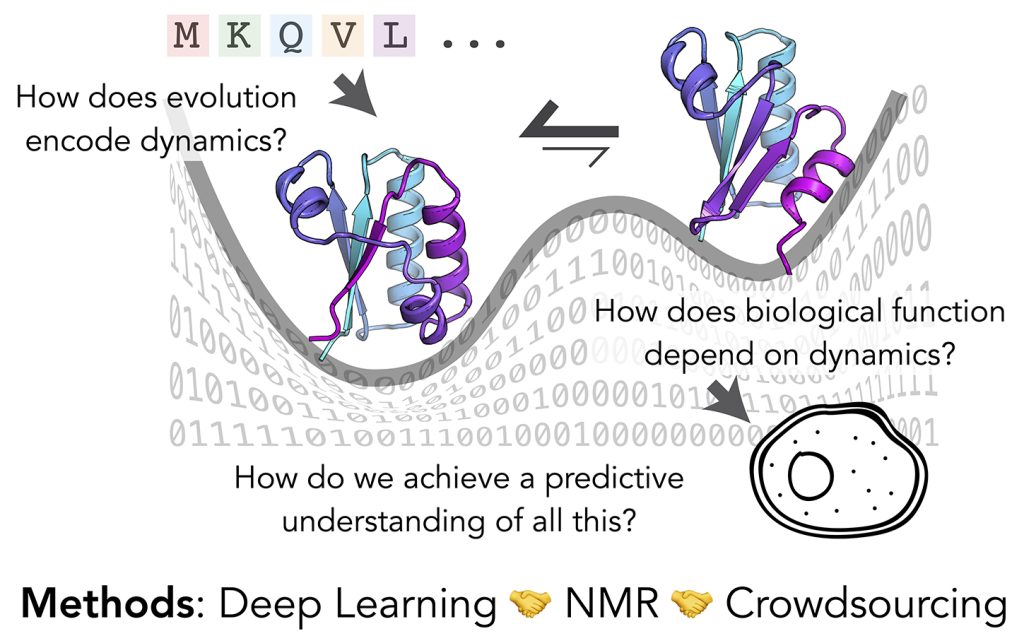
For life to exist as we know it, biomolecules must convert between distinct states with specific timing.
These dancing molecules are not just a fun quirk of biology – fundamental principles dictate that biomolecules
need kinetics for the exquisite specificity they achieve in the messy interior of a cell.
What could we unlock for fundamental molecular biology and its applications (drug discovery, enzyme design, and more)
if we had predictive power for the motions of biomolecules? The ability to accurately predict protein dynamics – multiple
conformations, their probabilities, and the kinetics of transitions between them – is the next grand challenge for
structural biology. My research seeks a quantitative and predictive understanding of biomolecular dynamics, and a deeper
understanding of how evolution shapes dynamics and function.
The Wayment-Steele lab will be at the interface of computational and experimental structural biology: we will formulate and
train models with principled biochemical questions in mind, and test their predictions with our own experiments.
We will initially focus on integrating deep learning and NMR dynamics experiments (relaxation dispersion), which are rich
in information and undervalued in protein deep learning, but currently limited in throughput. My postdoctoral research
focused on how AlphaFold and protein language models contain information on dynamics, yet deep learning in molecular biology
is evolving rapidly.
The fundamental principles my lab will focus on for any modeling approach are: how do we use sparse, yet multimodal data in
training; how do we construct meaningful prospective experimental tests, and how do we interpret what models are learning?
How will we maximize the impact of our results? De novo enzyme design will make for one stringent test in our own biological
systems. Being situated in the vibrant Biochemistry department at UW-Madison will also allow for numerous opportunities to apply
our work to more biologically relevant questions.
We will work with broader communities both at UW-Madison and beyond to conduct internet-scale crowdsourced challenges both for
deep learning and molecular design. This allows us to recruit diverse expertise and creativity to these challenges, and hopefully
allows many more worldwide to discover the delights of data on dancing molecules.
Predicting Biomolecular Dynamics
Understanding roles of dynamics in function, evolution, and design
Maximizing the Impact